I publish software so that other researchers can build upon my results and push the boundaries of science beyond where I have pushed them myself. My goal is for any trained computational researcher to be able to download my software and get it up and running. However, if you face some issues please let me know and I will help as best as I can. Open science is the best way forward.

How many sequencing reads are needed to detect a target T-Cell receptor using RNA sequencing? You can use this handy detection power calculator to find out. TCPower gives you the probability of detecting a target T-cell as a function of the number of sampled cells and sequencing reads. You can calibrate the calculator using your own pilot T-cell receptor sequencing data. See my article in Briefings in Bioinformatics for methodological details.
Implements a finite element modelling method for representing interstitial fibrosis (i.e. scarring) in the heart based on contrast enhanced cardiac MRI. The fibrosis is represented by cracks in the finite element mesh that are randomly created by the script. The local MRI image intensity, as well as some user-defined parameters control how many cracks are created. See my IEEE research paper for full details.

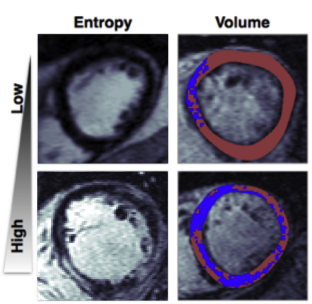
Calculates a set of metrics for quantifying the properties of heart tissue scars seen in contrast enhanced cardiac MRI. The metrics are related to the shape, size and texture, of the enhanced areas (i.e. scars). See my research article in JACC: Clinical Electrophysiology for an application to predicting future sudden cardiac arrest in patients with non-ischemic dilated cardiomyopathy.